biocurvature
| download: biocurvature_0112.rar |
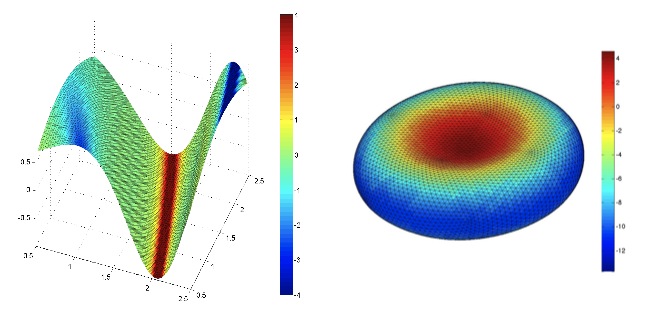
|
| 3D contour maps of the mean curvature of a sinusoidal membrane network (left) and a MD model of the red blood cell membrane (right). Refer to sections 3.1 and 3.3 of Fraternali et al., JOURNAL OF COMPUTATIONAL PHYSICS, 231, 528-540, 2012. |
Release notes for biocurvature
Developers:
© 2011-2012 All Rights Reserved
(*) corresponding author, f.fraternali@unisa.it
The “biocurvature” code performs the Local Maximum-Entropy (LME) regularization of a fluctuating membrane network, estimating the curvatures and in-plane strains of the mean shape of such a membrane.
The adopted LME scheme is fully described in the following reference:
Fraternali, F., Lorenz, C.D., Marcelli, G. On the estimation of the curvatures and bending rigidity of membrane networks via a local maximum-entropy approach. JOURNAL OF COMPUTATIONAL PHYSICS, 231, 528-540, 2012. ISSN: 0021-9991. DOI:10.1016/j.jcp.2011.09.017.
The compressed file biocurvature_0112.rar includes the following subfolders:
Animated example
Developers:
- main code: Fernando Fraternali(*)
- graphics: Fernando Fraternali and Vincenzo Ciancia
© 2011-2012 All Rights Reserved
(*) corresponding author, f.fraternali@unisa.it
The “biocurvature” code performs the Local Maximum-Entropy (LME) regularization of a fluctuating membrane network, estimating the curvatures and in-plane strains of the mean shape of such a membrane.
The adopted LME scheme is fully described in the following reference:
Fraternali, F., Lorenz, C.D., Marcelli, G. On the estimation of the curvatures and bending rigidity of membrane networks via a local maximum-entropy approach. JOURNAL OF COMPUTATIONAL PHYSICS, 231, 528-540, 2012. ISSN: 0021-9991. DOI:10.1016/j.jcp.2011.09.017.
The compressed file biocurvature_0112.rar includes the following subfolders:
- main: providing a read_me file; the executable biocurvature_01112.exe for windows; the associated Matlab plotting code biocurvature_plot_0112.m; the Matlab library MyCrustOpen (needed for plotting purposes); and the current release notes;
- examples: providing input/output files for the illustrative examples described in sections 3.1 (sinusoidal membrane) and 3.3 (red blood cell membrane) of Fraternali et al., JCOMP 2011;
- papers: providing some reference papers about the LME regularization scheme; the given demonstrative examples and related topics (limiting elastic energies and elastic coefficients of fluctuating membrane networks).
Animated example
Area stretching fluctuations of a molecular dynamics model of the red blood cell membrane
(bibliographic reference)